Hi, there! This is Lihua Wu.
I'm best known as a Biologist, a passionate Bioinformatician and Data lover, and a dedicated science educator.
Learn about what I doI'm best known as a Biologist, a passionate Bioinformatician and Data lover, and a dedicated science educator.
Learn about what I doBiologist, Bioinformatician, Datamania and Science educator
I have more than 10 years of research experience in plant reproduction and plant adaptation to cold temeperature. I have accquired extensive skills in molecular biology, biochemistry, genetics and genomics.
I have been working with large biological datasets for more that 5 years, such as RNA-seq, Mi-seq and PacBio data. I am familiar with many Bioinformatics software and tools. I have extensive experience in NGS sequence analysis and data visualization.
I have been exposed to many aspects of data science. I am always keeping updated with new skills in Statistics, machine learning and artificial intelligence.
I am passionate about science education and outreach.
I have extensive experience in biology research. My focus is molecular biology, genetics, biochemistry and genomics.
I did some early data exploration of the 2019-nCov to help visualize the current outbreak status.
One of my Ph.D. projects is sequencing, assembly and analysis of the highly-repetitive S-locus region of Petunia inflata. My contribution involves preparing sample for Illumina Miseq sequencing and PacBio RSII sequencing and sequence analysis and visualization.
One of my Ph.D. projects is investigating key amino acid residues involved in specific interaction between S-RNase proteins and S-locus F-box proteins. My contribution involves molecular cloning, plant transformation, in vivo function analysis, molecular modeling and docking.
My master study focused on plant adaptation to low temperature. I used Nicotiana tabaccum and Arabidopsis thaliana as model plants to study the mechanism of plant cold resistance. I first-authored and/or co-authored more than 10 peer-reviewed publications.
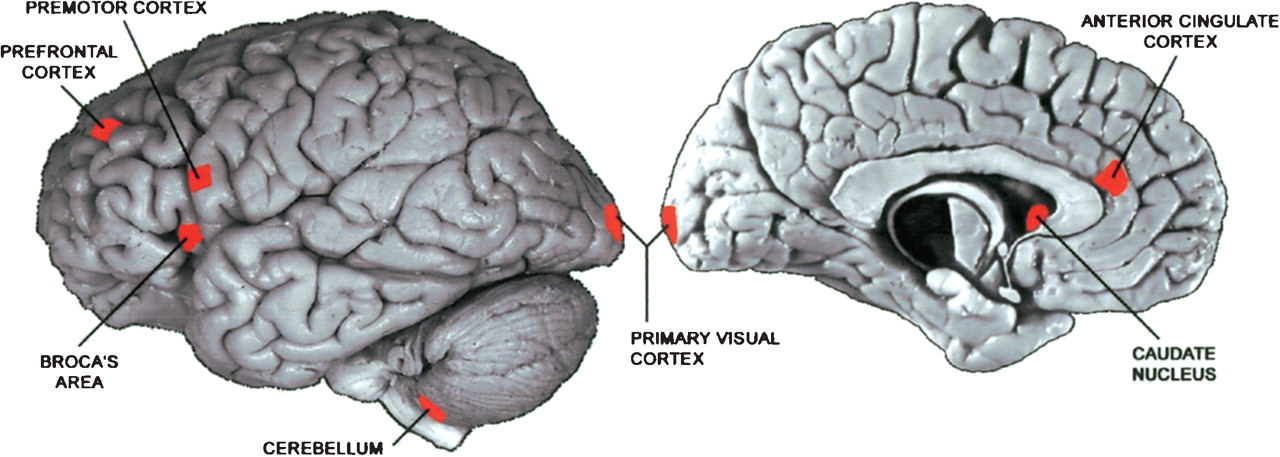
Khaitovich et al (2004) compared gene expression in 7 homologous regions of human and chimpanzee brains. There were 3 human brains and 3 chimpanzee brains available for the study. Each brain was dissected to obtain tissue samples for each of the 7 regions.
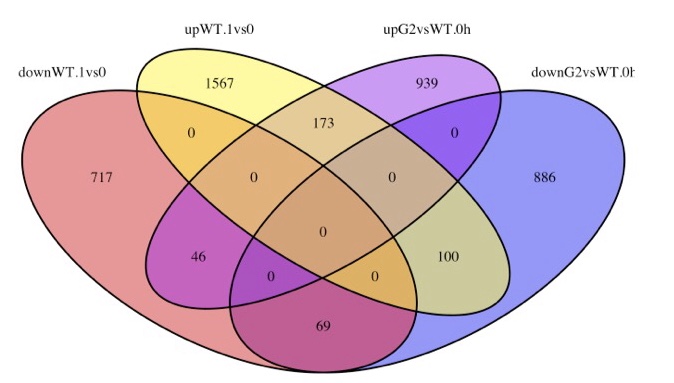
In this project, I used edgeR and voom to analyze Arabidopsis RNA-seq data. I identified similar number of differentially expressed genes with the author. I use AgriGO and Ami GO2 online tools to do GO enrichment analysis, and observe similar significant GO terms with the author.
Another interesting project I worked on during my Ph.D is Subcellular Localization and Temporal Expression of S2-SLF1 as Revealed by Confocal Microscopic Image Analysis Using S2-SLF1-(super-folder)GFP and/or Pollen-Specific Organelle Markers
I genuinely enjoy teaching students in classrooms, labs and field trips. I have taught Introductory Plan Biology to high school students in a row of four years using self-developed curriculum, which consists of a serie of lectures, a research project and field trips. I have taught Molecular Biology, Microbiology and Biochemistry lectures and labs to undergraduate students for a total of 7 times during my graduate study.
Download Lihua's Teaching Statement